Reconstructing the trophic radiation of Lake Victoria Cichlids
Lake Victoria is the largest lake in Africa and harbored more than 500 endemic species of haplochromine cichlid fish. The lake not only was at a low stand but dried up completely during the Late Pleistocene, approx. 15000 years before present (Seehausen 2006). These results imply that the rate of speciation of cichlid fish in this tropical lake has been extremely rapid! However, only little is know about the exact order of emergence and respective speciation rates of the different trophic guilds (McGee et al 2020). Using the temporal information from the sediment cores, as well as other data about the ancient lake communities, we are hoping to learn more about this adaptive radiation, which is among one of the most rapid cases of adaptive evolution.
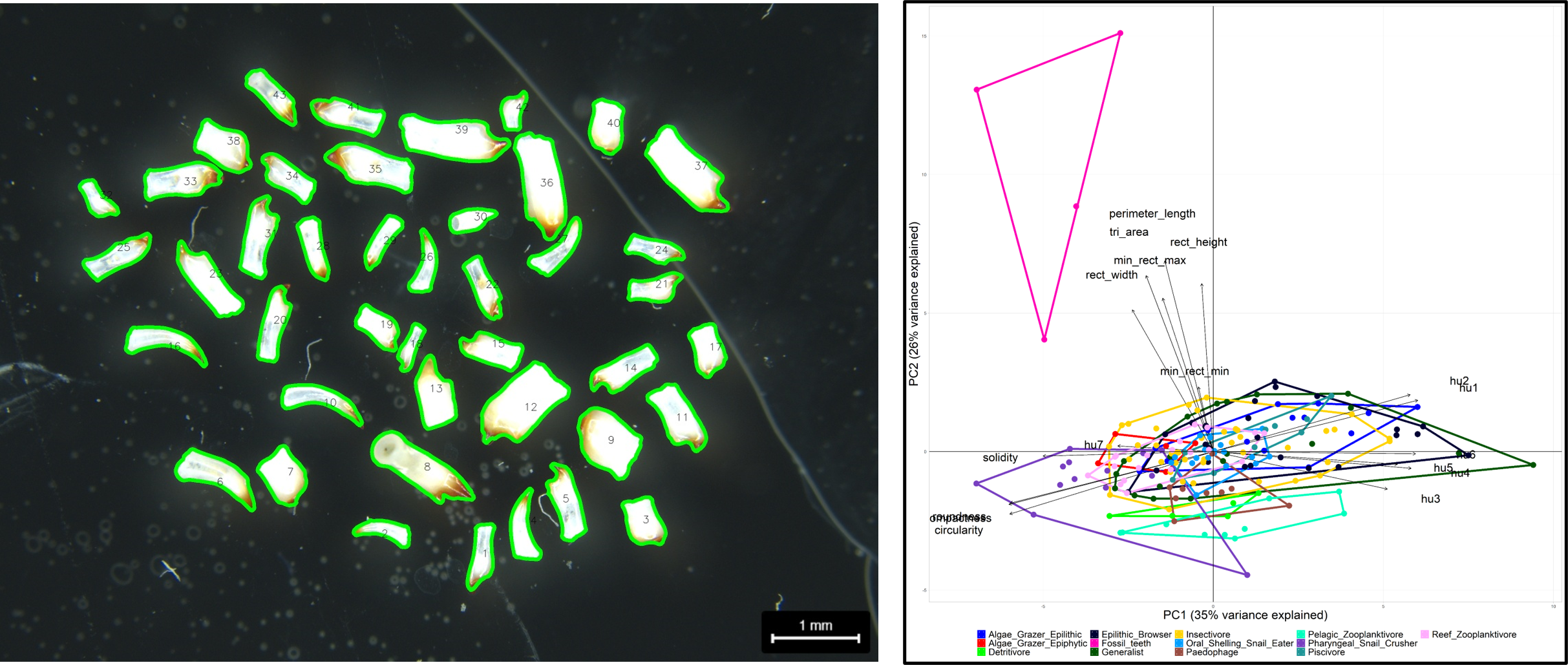
In a first attempt I used phenopype to segment 2D-representations of teeth from contemporary cichlid species to construct a classifier would map fossil teeth found in sediment cores onto contemporary trophic guilds. However, on the hand we did not have enough material to create a robust classifier, and on the other hand, this was a destructive procedure where valuable cichlid specimens would have had to be sacrificed to retrieve their teeth. We therefore turned to computer tomography as a non-invasive imaging technique.
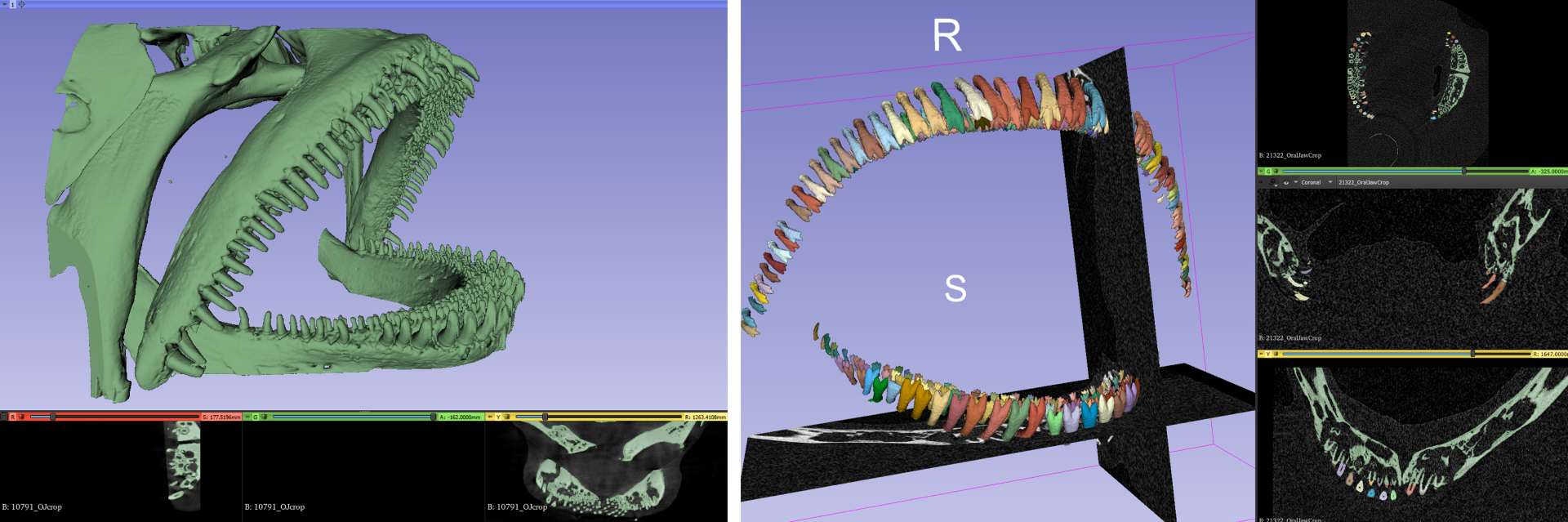
For the obtained 3D-images the goal is the same: segment the foreground (individual teeth in both jaws and all tooth-rows) from the background (the fish jaws and skull). We have started to use biomedisa (Lösel et al. 2020) for automatic segmentation of shapes in 3D images - this work is currently ongoing.
References
Lösel, P. D., van de Kamp, T., Jayme, A., Ershov, A., Faragó, T., Pichler, O., Jerome, N. T., Aadepu, N., Bremer, S., Chilingaryan, S. A., Heethoff, M., Kopmann, A., Odar, J., Schmelzle, S., Zuber, M., Wittbrodt, J., Baumbach, T., & Heuveline, V. (2020). Introducing Biomedisa as an open-source online platform for biomedical image segmentation. Nature Communications, 11(1), 1–14.
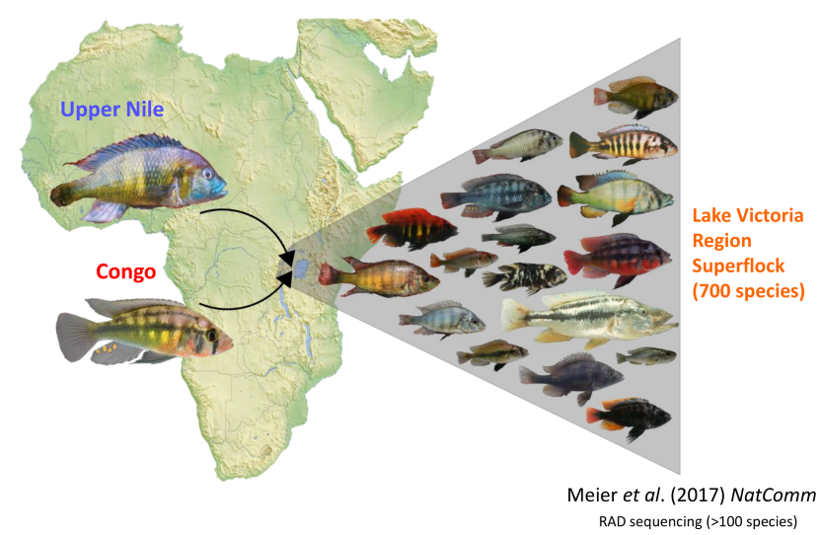
Meier, J. I., D. A. Marques, S. Mwaiko, C. E. Wagner, L. Excoffier, and O. Seehausen. 2017. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nature communications 8:14363.
Seehausen, O. 2006. African cichlid fish: a model system in adaptive radiation research. Proceedings. Biological sciences / The Royal Society 273:1987–1998.
McGee, M. D., S. R. Borstein, J. I. Meier, D. A. Marques, S. Mwaiko, A. Taabu, M. A. Kishe, B. O’Meara, R. Bruggmann, L. Excoffier, and O. Seehausen. 2020. The ecological and genomic basis of explosive adaptive radiation. Nature 586:75–79.